Community developed variant calling pipelines
Brad Chapman
Bioinformatics Core, Harvard School of Public Health
@chapmanb
20 July 2013
Challenges
Complex, rapidly changing pipelines

Large number of specialized dependencies
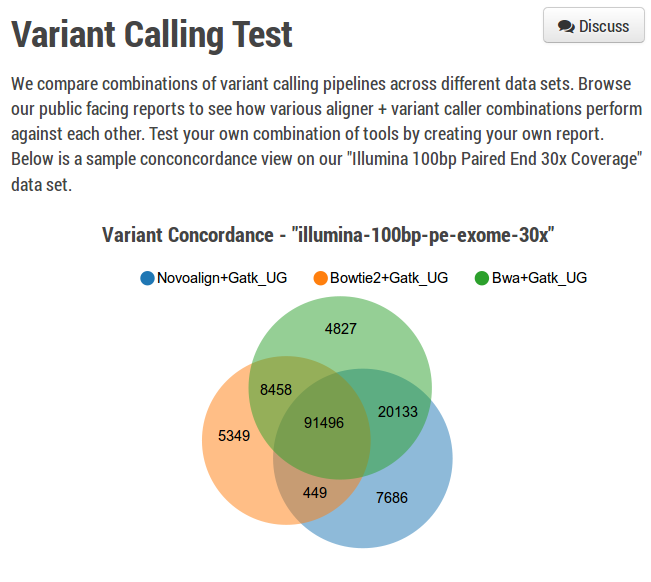
Quality differences between calling methods
Scaling on full ecosystem of clusters

Solution

http://www.amazon.com/Community-Structure-Belonging-Peter-Block/dp/1605092770
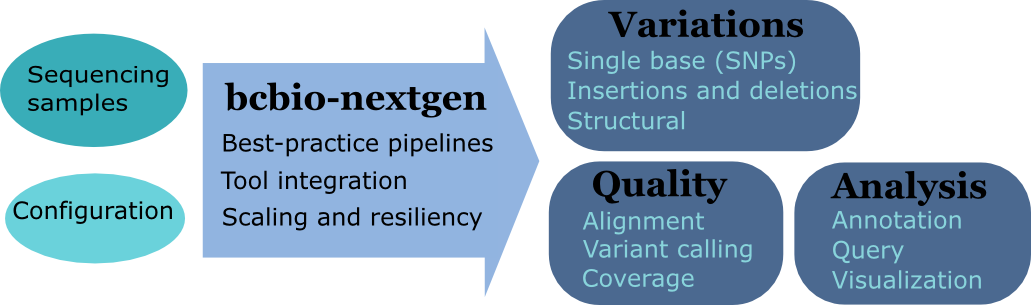
Development goals
- Quantifiable
- Analyzable
- Scalable
- Reproducible
- Community developed
- Accessible
Quantify quality
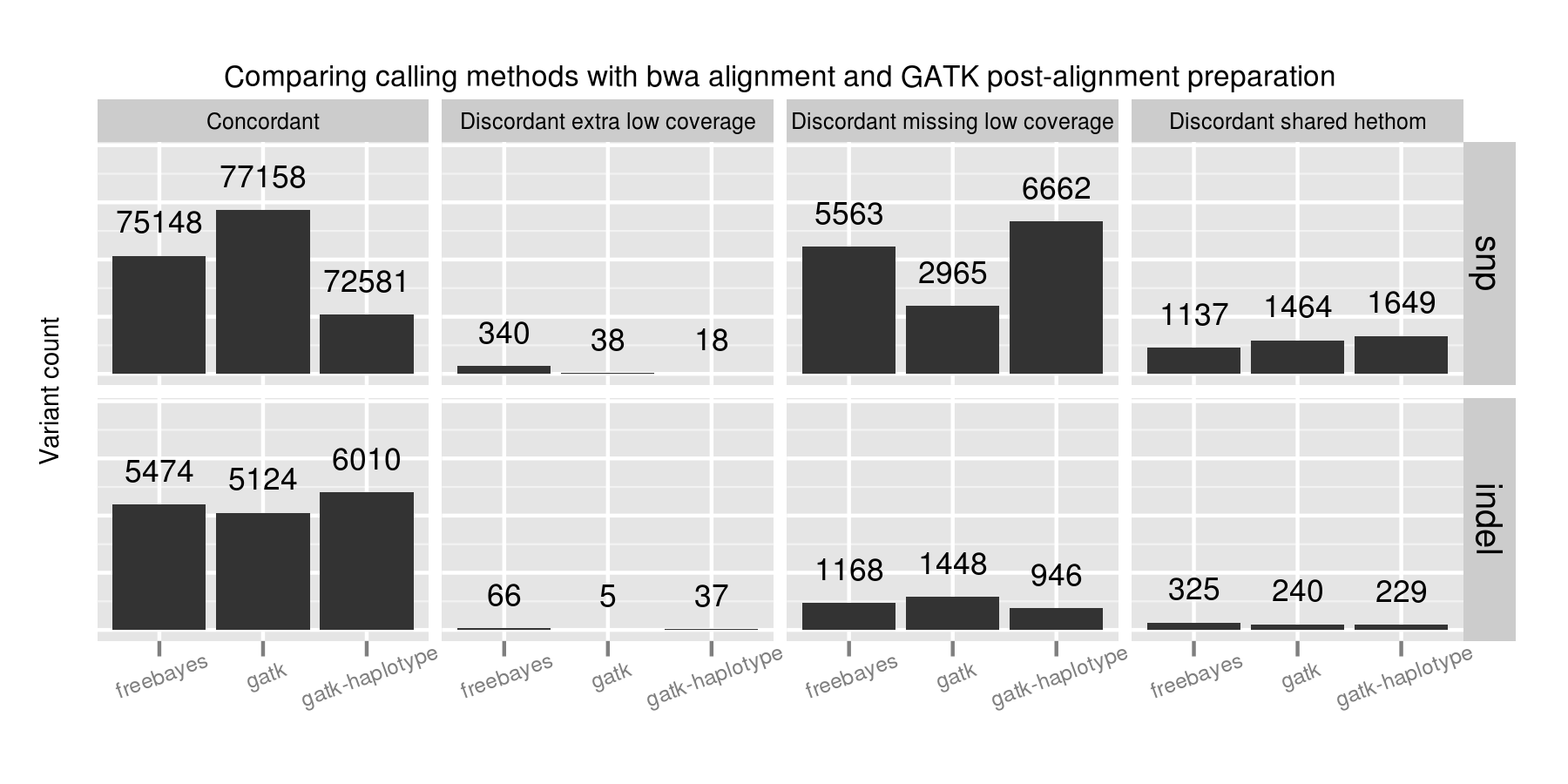
- Reference materials: http://www.genomeinabottle.org/
- Quantification details: http://j.mp/bcbioeval
Known unknowns
- Coverage: summarize what you can't assess
- Structural: large, complex rearrangements
Analysis
Query
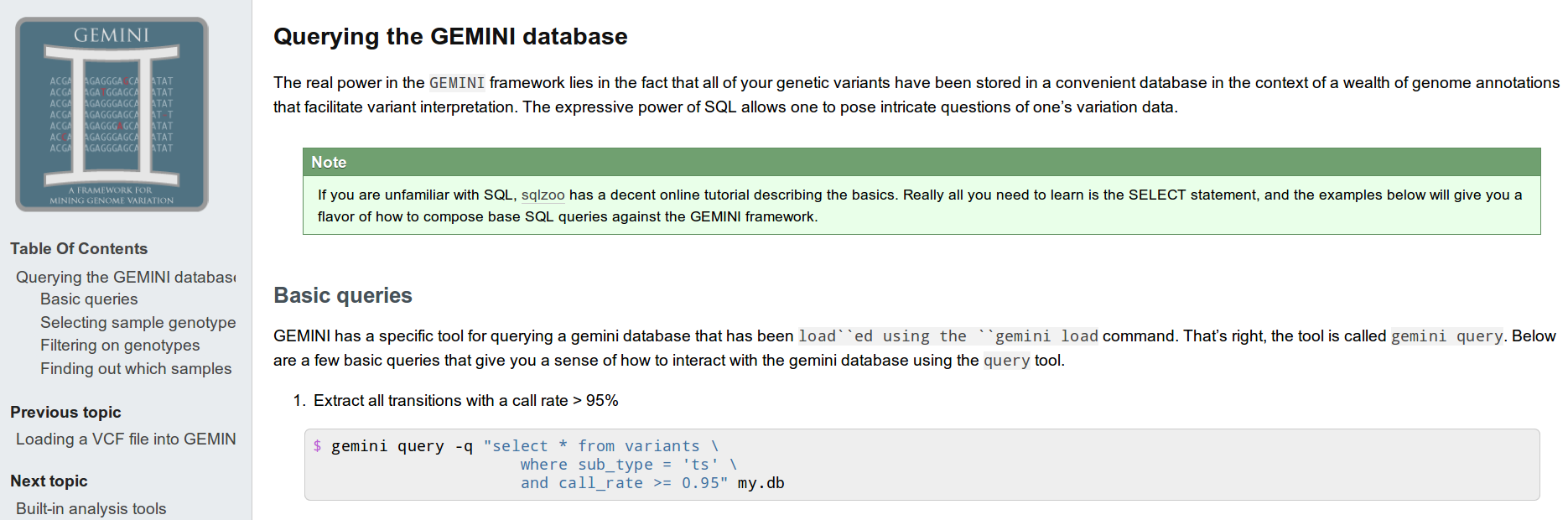
Visualize
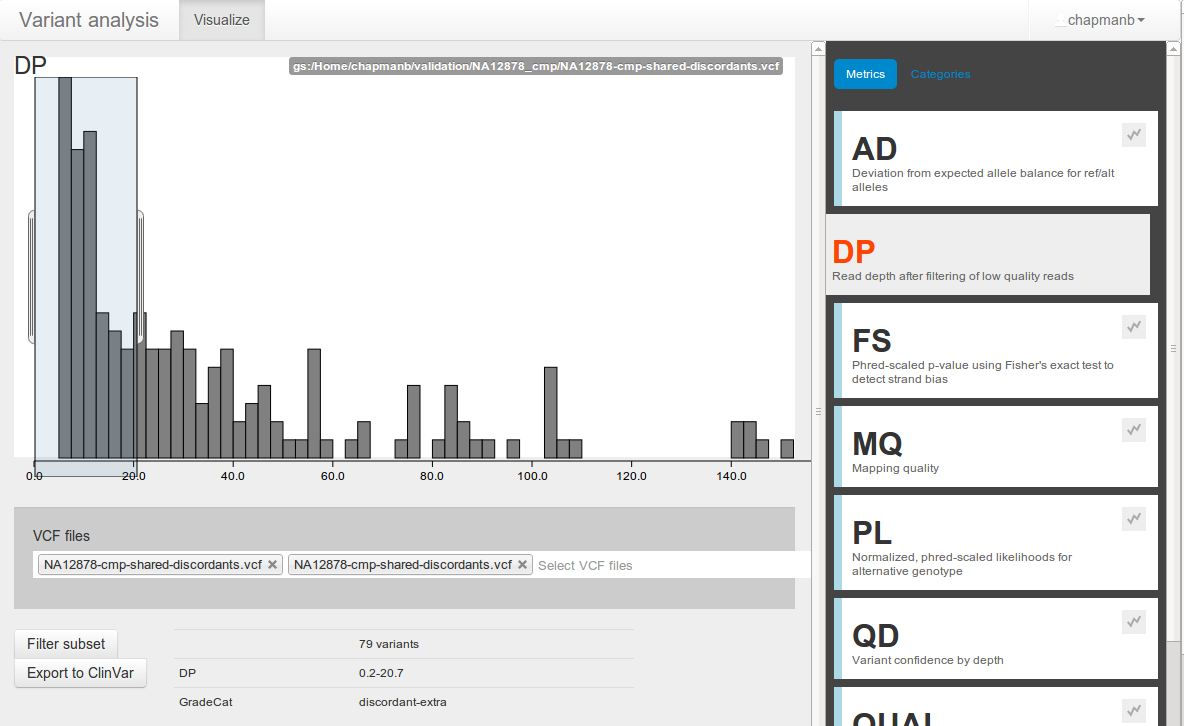
Parallel scaling
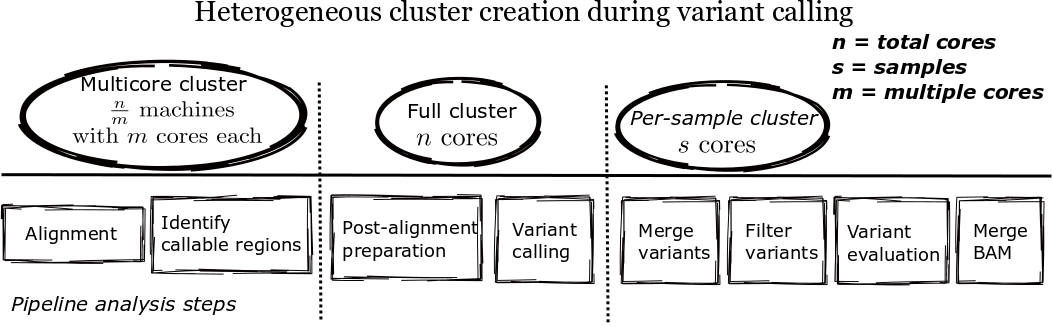
- Infrastructure details: http://j.mp/bcbioscale
- IPython: http://ipython.org/ipython-doc/dev/parallel/index.html
Better parallel blocks
Reproducibility
- Express intentions at a high level
- Revision controlled configuration
- Handle complex distributed logging
- Provenance tracking
Configuration
- files: [NA12878-NGv3-LAB1360-A_1.fastq.gz, NA12878-NGv3-LAB1360-A_2.fastq.gz] description: NA12878 analysis: variant2 genome_build: GRCh37 algorithm: aligner: bwa recalibrate: gatk realign: gatk variantcaller: [gatk, freebayes, gatk-haplotype] coverage_interval: exome coverage_depth: high platform: illumina quality_format: Standard validate: NA12878-nist-v2_13-NGv3-pass.vcf
Provenance
- Excellent logging
- Third party version tracking
- Beyond logging:
- BioLite: https://bitbucket.org/caseywdunn/biolite
- Arvados: https://arvados.org/
Community developed
- Fully automated installation: CloudBioLinux
- Deployable on multiple clusters (LSF, SGE, Torque…)
- API for new aligners and variant callers
- Open source, hackable and documented
Automated installation
- Single biggest software problem: running for the first time
- Bootstrap from bare machine to ready-to-go pipeline
- Builds off existing installation work: CloudBioLinux
- Provide example pipelines with real data
Accessible

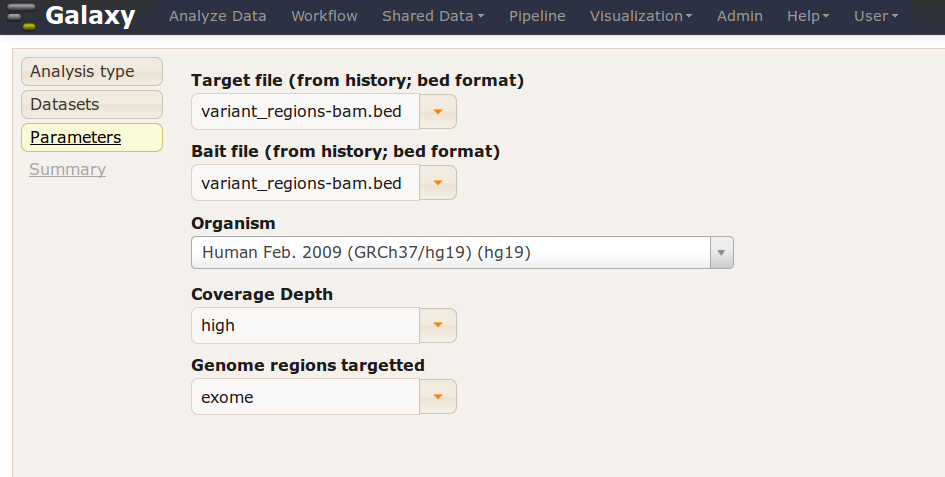
Galaxy
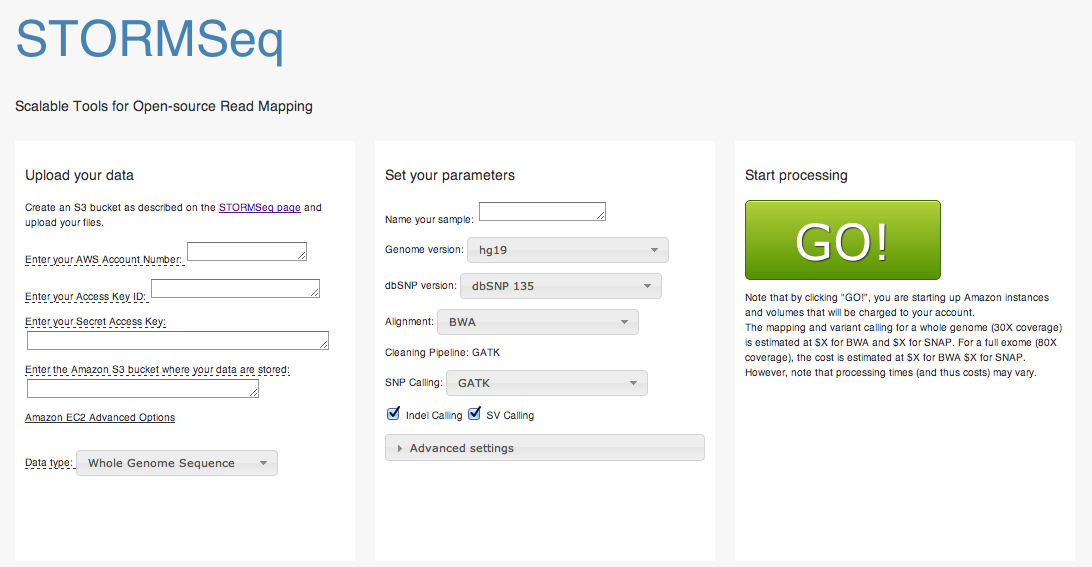
STORMSeq
Summary
- Community developed pipelines > challenges
- Focus
- Assessing quality: good science
- Analysis: enable exploration
- Scalability: finish in time
- Reproducibility: show your work
- Widely accessible